2023
Hattori M., Horigome C., Aspert Théo, Charvin G., Kobayashi T.
In this collaborative work, I have shared a microfluidic device to track single yeast cells throughout aging, trained my collaborators to use it, and reviewed the biological & scientific content of the study.
Genes & Genetic Systems, (2023) - DOI: 10.1266/ggs.22-00100
AbstractGenome instability is a major cause of aging. In the budding yeast Saccharomyces cerevisiae, instability of the ribosomal RNA gene repeat (rDNA) is known to shorten replicative lifespan. In yeast, rDNA instability in an aging cell is associated with accumulation of extrachromosomal rDNA circles (ERCs) which titrate factors critical for lifespan maintenance. ERC accumulation is not detected in mammalian cells, where aging is linked to DNA damage. To distinguish effects of DNA damage from those of ERC accumulation on senescence, we re-analyzed a yeast strain with a replication initiation defect in the rDNA, which limits ERC multiplication. In aging cells of this strain (rARS-∆3) rDNA became unstable, like in wild-type cells, whereas ERCs were much less accumulated. Single cell aging analysis revealed that rARS-∆3 cells follow a linear survival curve and can have a wild-type replicative lifespan, although a fraction of cells stopped dividing earlier than wild type. Doubling times of rARS-∆3 cells appear to increase in the final cell divisions. Our results suggest that senescence in rARS-∆3 is linked to the accumulation of DNA damage like in mammalian cells, rather than to elevated ERC levels. Therefore, this strain could be a good model system to study damage-induced aging.
2022
Aspert Théo, Charvin Gilles
HardwareX (In press), (2022) - DOI: Coming soon
bioRxiv, (2022) - DOI: 10.1101/2022.11.15.516605
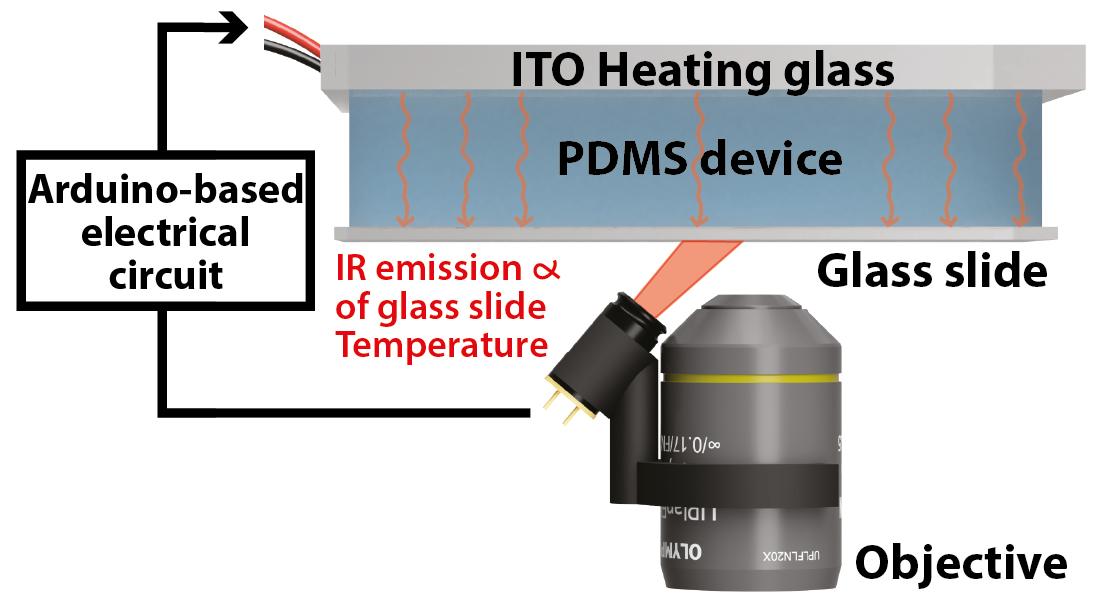
AbstractMicrofluidic systems are widely used in biology for their ability to control environmental parameters. Specifically, cell culture or chemistry in microfluidic devices requires tight control of the temperature. In addition, microfluidic devices can be made transparent to visible light and compatible with inverted microscopes. Yet, the current temperature control systems that allow high-resolution microscopy either require a set of complex secondary channels, a bulky, expensive, and microscope-dependent incubator, or fail to produce a homogenous temperature profile across the sample area. Here, we present HeatChips, a simple, cost-effective system to heat samples inside PDMS-based microfluidic devices in a homogeneous manner. It is based on a transparent heating glass in contact with the top of the microfluidic device, and a contactless, infrared temperature sensor attached to the objective that directly reads the temperature of the bottom of the chip. This portable system is compatible with most chip designs and allows imaging of the sample on inverted microscopes for extended periods of time without any optical restriction, for a cost of less than 100€.
Quintin Sophie, Aspert Théo, Charvin Gilles
In this collaborative work, I have engineered a microfluidic device to trap C. elegans worm for several hours while they are exposed to stress and imaged at very high speeds.
PLoS ONE, (2022) - DOI: 10.1371/journal.pone.0274226
bioRxiv, (2021) - DOI: 10.1101/2021.07.26.451501
AbstractEnvironmental oxidative stress threatens cellular integrity and should therefore be avoided by living organisms. Yet, relatively little is known about environmental oxidative stress perception. Here, using microfluidics, we showed that like I2 pharyngeal neurons, the tail phasmid PHA neurons function as oxidative stress sensing neurons in C. elegans, but display different responses to H2O2 and light. We uncovered that different but related receptors, GUR-3 and LITE-1, mediate H2O2 signaling in I2 and PHA neurons. Still, the peroxiredoxin PRDX-2 is essential for both, and might promote H2O2-mediated receptor activation. Our work demonstrates that C. elegans can sense a broad range of oxidative stressors using partially distinct H2O2 signaling pathways in head and tail sensillae, and paves the way for further understanding of how the integration of these inputs translates into the appropriate behavior.

Aspert Théo, Hentsch Didier, Charvin Gilles
eLife, (2022) - DOI: 10.7554/eLife.79519
bioRxiv, (2021) - DOI: 10.1101/2021.10.05.463175
AbstractAutomating the extraction of meaningful temporal information from sequences of microscopy images represents a major challenge to characterize dynamical biological processes. So far, strong limitations in the ability to quantitatively analyze single-cell trajectories have prevented large-scale investigations to assess the dynamics of entry into replicative senescence in yeast. Here, we have developed DetecDiv, a microfluidic-based image acquisition platform combined with deep learning-based software for high-throughput single-cell division tracking. We show that DetecDiv can automatically reconstruct cellular replicative lifespans with high accuracy and performs similarly with various imaging platforms and geometries of microfluidic traps. In addition, this methodology provides comprehensive temporal cellular metrics using time-series classification and image semantic segmentation. Last, we show that this method can be further applied to automatically quantify the dynamics of cellular adaptation and real-time cell survival upon exposure to environmental stress. Hence, this methodology provides an all-in-one toolbox for high-throughput phenotyping for cell cycle, stress response, and replicative lifespan assays.
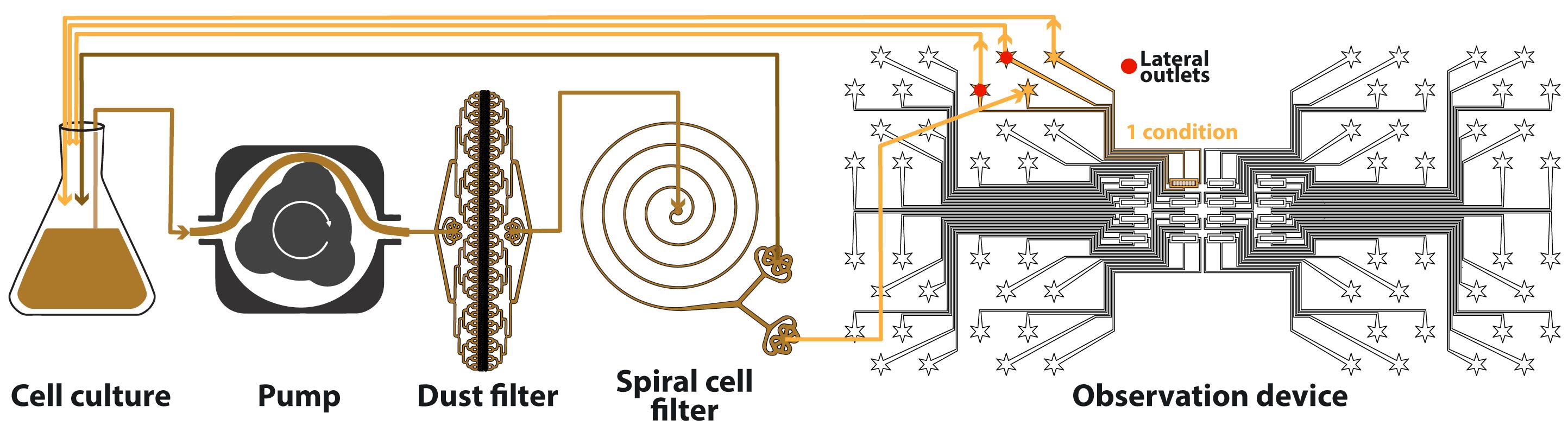
Aspert Théo, Jacquel Basile, Gilles Charvin
Bioprotocols, (2022) - DOI: 10.21769/BioProtoc.4470
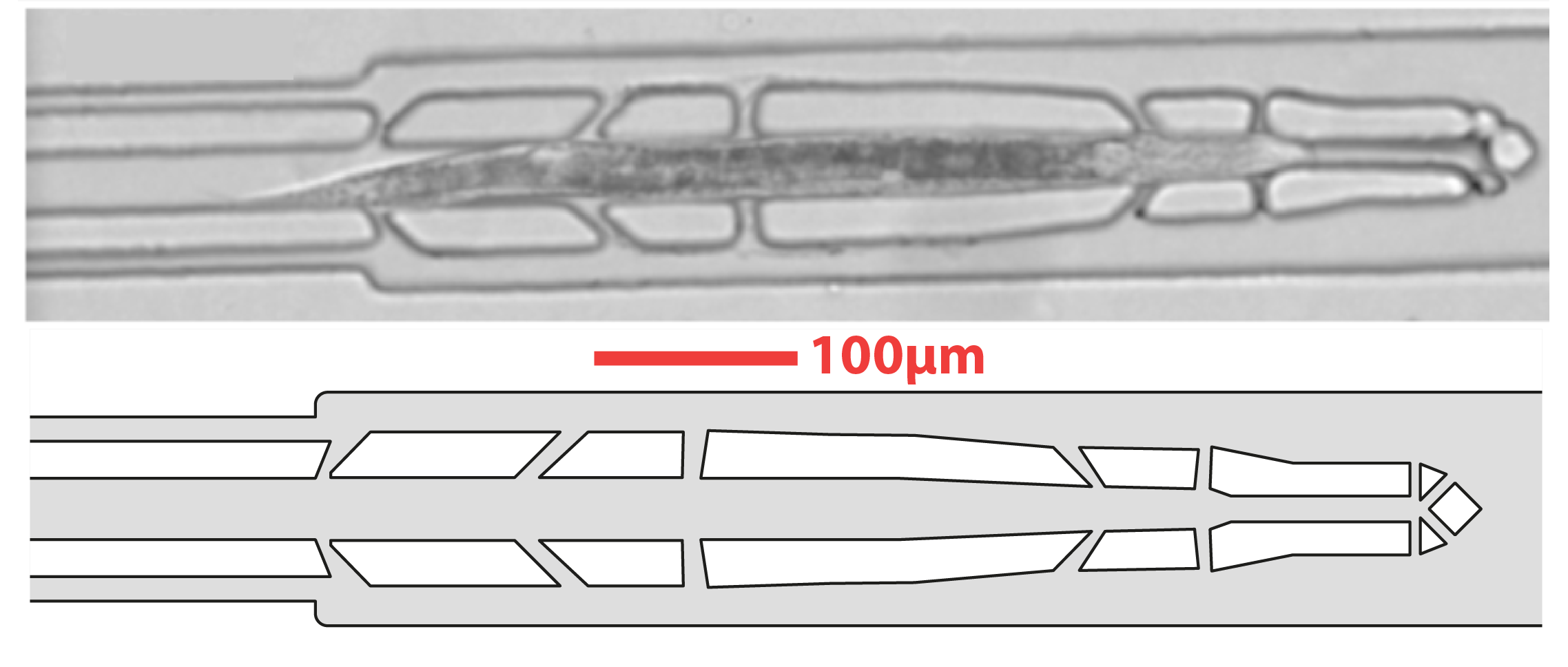
AbstractMicroorganisms have evolved adaptive strategies to respond to the autonomous degradation of their environment. Indeed, a growing culture progressively exhausts nutrients from its media and modifies its composition, but how single cells react to these modifications remains difficult to study since it requires population-scale growth experiments to allow cell proliferation to have a collective impact on the environment, while monitoring the same individuals for days. For this purpose, we have previously described an integrated microfluidic pipeline, based on continuous separation of the cells from the media and subsequent perfusion of the filtered media in an observation chamber containing isolated single-cells. Here, we provide a detailed protocol to implement this methodology, including the setting up of the microfluidic system and the processing of timelapse images.
Anne C. Meinema, Aspert Théo, Sung Sik Lee, Gilles Charvin, Yves Barral
In this collaborative work, I have conceived a microfluidic device to track yeast cells during the process of aging, participated in the development of the experimental pipeline, ran some microfluidics experiments and reviewed the biological & scientific content of the study.
eLife, (2022) - DOI: 10.7554/eLife.71196
bioRxiv, (2021) - DOI: 10.1101/2021.06.28.450139
AbstractThe nuclear pore complex (NPC) mediates nearly all exchanges between nucleus and cytoplasm, and in many species it changes composition as the organism ages. However, how these changes arise and whether they contribute themselves to ageing is poorly understood. We show that SAGA-dependent attachment of DNA circles to NPCs in replicatively ageing yeast cells causes NPCs to lose their nuclear basket and cytoplasmic complexes. These NPCs were not recognized as defective by the NPC quality control machinery (SINC) and not targeted by ESCRTs. They interacted normally or more effectively with protein import and export factors but specifically lost mRNA export factors. Acetylation of Nup60 drove the displacement of basket and cytoplasmic complexes from circle-bound NPCs. Mutations preventing this remodeling extended the replicative lifespan of the cells. Thus, our data suggest that the anchorage of accumulating circles locks NPCs in a specialized state and that this process is intrinsically linked to the mechanisms by which ERCs promote ageing.
2021
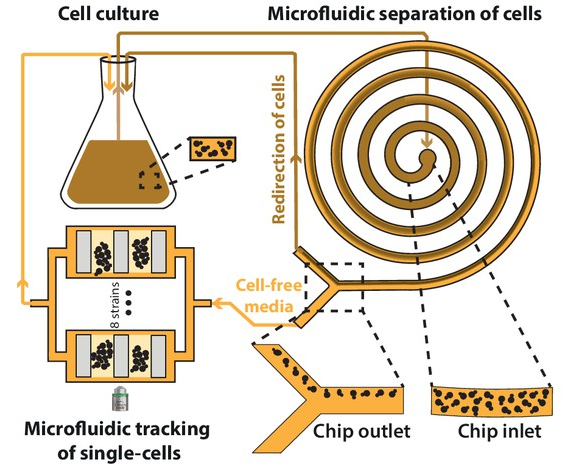
Aspert Théo*, Jacquel Basile*, Laporte Damien, Sagot Isabelle, Charvin Gilles
* Equally contributed to this work
eLife, (2021) - DOI: 10.7554/eLife.73186
AbstractThe life cycle of microorganisms is associated with dynamic metabolic transitions and complex cellular responses. In yeast, how metabolic signals control the progressive choreography of structural reorganizations observed in quiescent cells during a natural life cycle remains unclear. We have developed an integrated microfluidic device to address this question, enabling continuous single-cell tracking in a batch culture experiencing unperturbed nutrient exhaustion to unravel the coordination between metabolic and structural transitions within cells. Our technique reveals an abrupt fate divergence in the population, whereby a fraction of cells is unable to transition to respiratory metabolism and undergoes a reversible entry into a quiescence-like state leading to premature cell death. Further observations reveal that non-monotonous internal pH fluctuations in respiration-competent cells orchestrate the successive waves of protein super-assemblies formation that accompany the entry into a bona fide quiescent state. This ultimately leads to an abrupt cytosolic glass transition that occurs stochastically long after proliferation cessation. This new experimental framework provides a unique way to track single-cell fate dynamics over a long timescale in a population of cells that continuously modify their ecological niche.
2019
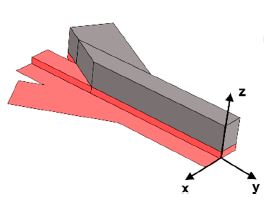
Constantinou I*, Jendrusch M*, Aspert Théo, Görlitz F, Schulze A, Charvin G, Knop M.
* Equally contributed to the work
In this collaborative work, I have designed and fabricated a microfluidic device to focus cells in 3D in order to reproduce a cytometer-on-chip. I also participated in the development of a machine learning algorithm to classify the resulting images of cells into cell types.
Micromachines, (2019) - DOI: 10.3390/mi10050311
AbstractSingle-cell analysis commonly requires the confinement of cell suspensions in an analysis chamber or the precise positioning of single cells in small channels. Hydrodynamic flow focusing has been broadly utilized to achieve stream confinement in microchannels for such applications. As imaging flow cytometry gains popularity, the need for imaging-compatible microfluidic devices that allow for precise confinement of single cells in small volumes becomes increasingly important. At the same time, high-throughput single-cell imaging of cell populations produces vast amounts of complex data, which gives rise to the need for versatile algorithms for image analysis. In this work, we present a microfluidics-based platform for single-cell imaging in-flow and subsequent image analysis using variational autoencoders for unsupervised characterization of cellular mixtures. We use simple and robust Y-shaped microfluidic devices and demonstrate precise 3D particle confinement towards the microscope slide for high-resolution imaging. To demonstrate applicability, we use these devices to confine heterogeneous mixtures of yeast species, brightfield-image them in-flow and demonstrate fully unsupervised, as well as few-shot classification of single-cell images with 88% accuracy.
BaCell3D, building advanced multiCellular systems in 3D Link
Capturing the first event of the aging cascade using microfluidics, high-throughput microscopy and deep-learning
PhD Prize ceremony of the Society of Biology of Strasbourg - Strasbourg, April 2023
Measuring single-cells dynamics using microfluidics, high-throughput microscopy and deep-learning
GDR Micro-nano-fluidics - Annual meeting - Lyon, April 2023
GDR Organoids, Annual meeting - Paris, December 2022, Link
Measuring single-cells dynamics using microfluidics, imaging and deep-learning
Institut Pasteur internal seminar - Invited by the Baroud's lab - Paris (France), November 2022
Microfluidics applications: from research to drug discovery, a workshop organized by the Drug Institute of Strasbourg (IMS) - November 2022, Link
French Society for Stem Cell Research - 5th Annual Meeting - October 2022, Link
EMBL Microfluidics - Heidelberg (Germany), July 2022, Link
LMO 14th International Conference - October 2021, Link
Tracking single-cells during aging and environmental adaptation using microfluidics, high-throughput microscopy and deep-learning
GDR Micro-nano-fluidics - Annual meeting - Toulouse, September 2021
Replicative aging: capturing the start of the countdown to death using high-throughput microfluidics and deep-learning
IGBMC Seminar Series - June 2021
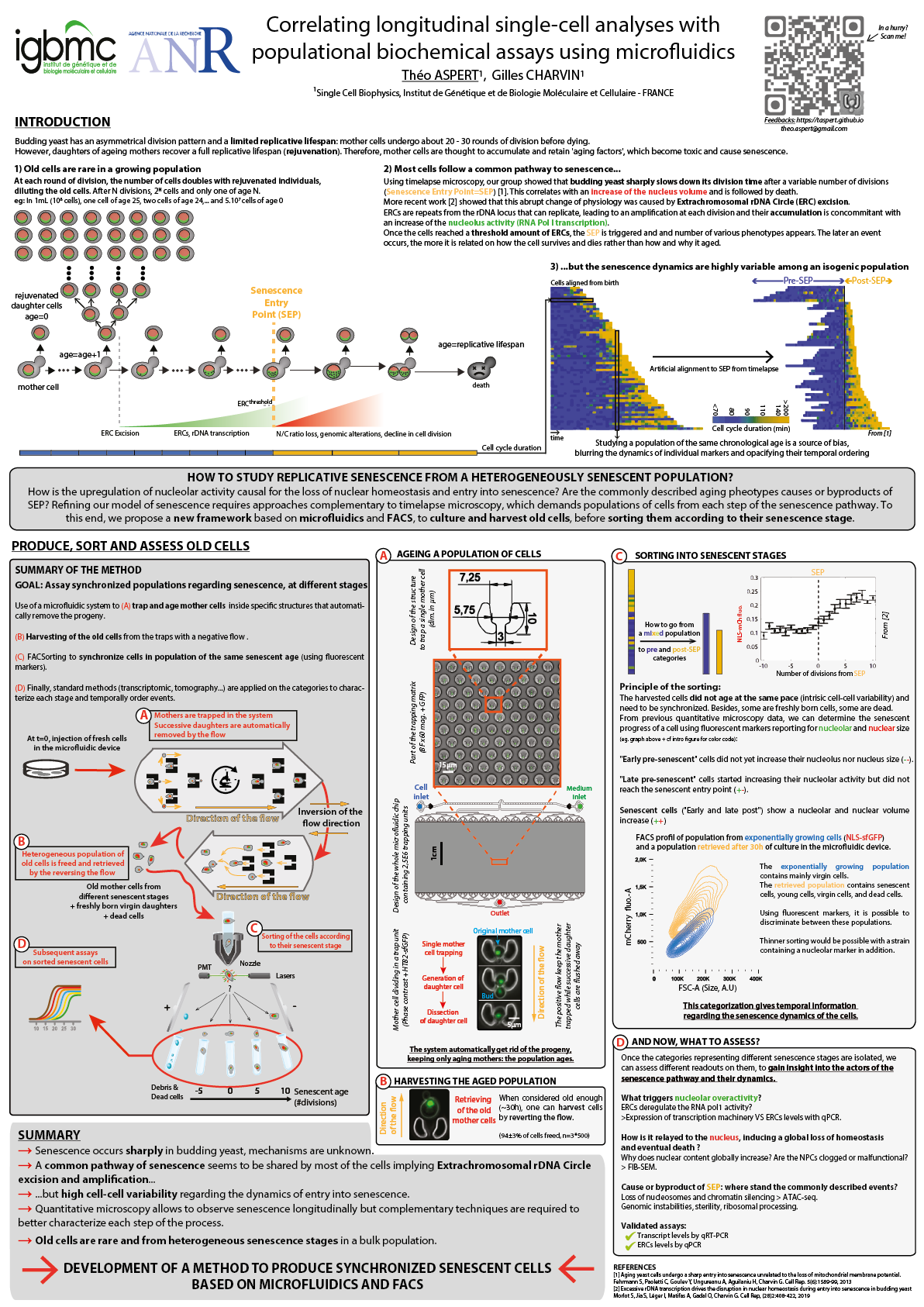
Correlating longitudinal single-cell analyses with populational biochemical assays using microfluidics
International Conference on Yeast Genetics and Molecular Biology - Gothenburg (Sweden), August 2019
High-throughput harvesting of replicative aged cells using microfluidics
High-Throughput Technologies Applied to Yeasts - Strasbourg (France), November 2018
Selective harvesting of replicative aged cells using microfluidics
Experimental Approaches to Evolution and Ecology Using Yeast and Other Model Systems - EMBL - Heidelberg (Germany), October 2018